back to index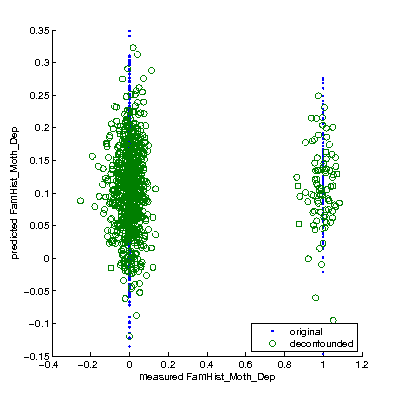
Correlation/prediction results for subject measure 43 (FamHist_Moth_Dep)
815 subjects had a valid FamHist_Moth_Dep measure.
Multivariate prediction (GLM-based, automatic feature selection, leave-one-family-out prediction)
Original data space: r=0.06 CoD=-0.04 Deconfounded space: r=0.03 CoD=-0.03
Scatterplot shows predicted-FamHist_Moth_Dep vs measured-FamHist_Moth_Dep (in original and deconfounded data space).
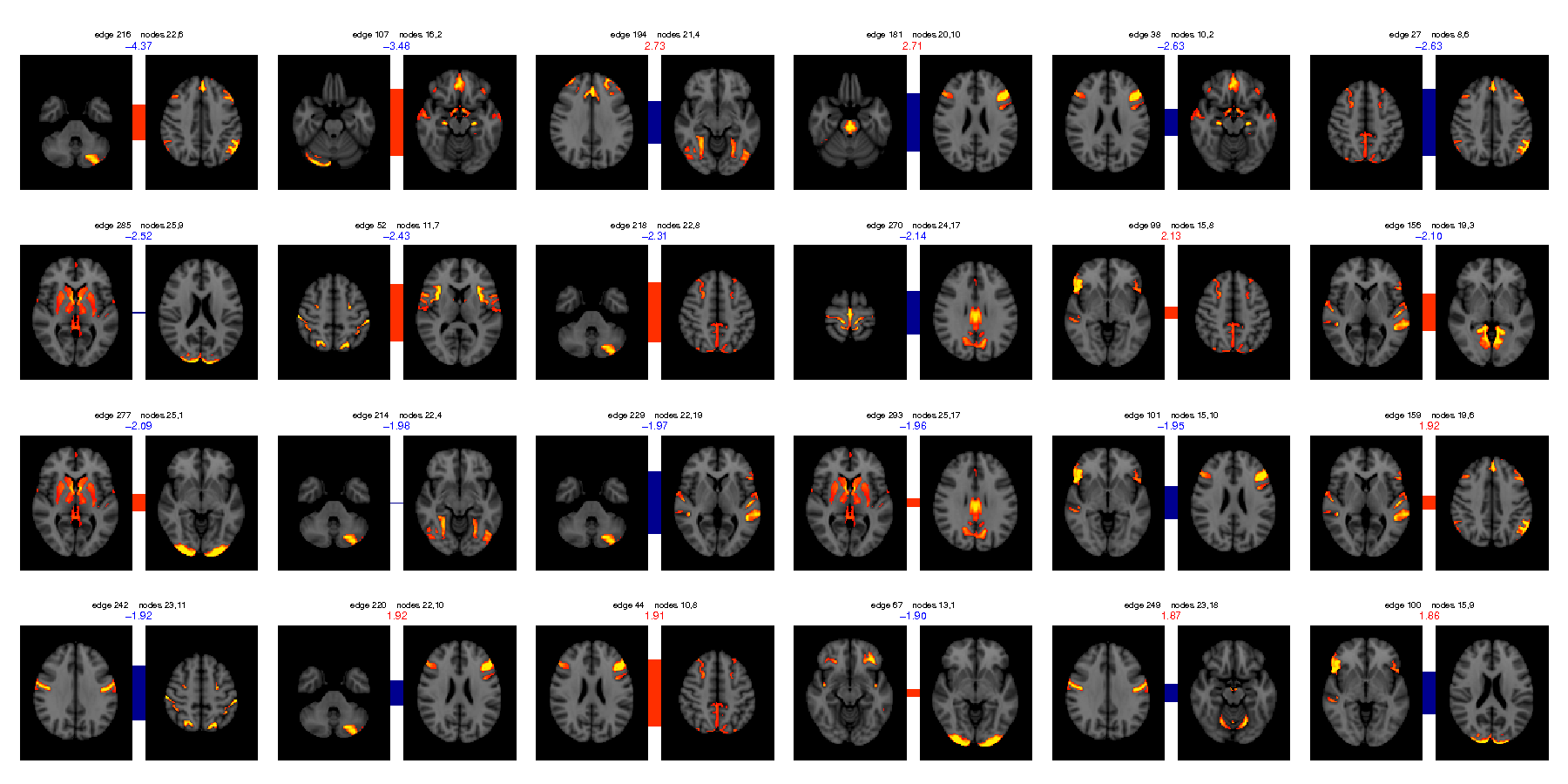
Univariate regression (regressing each netmat element independently against FamHist_Moth_Dep, correcting for multiple comparisons across elements, using PALM permutation testing, taking into account family structure).
Number of significantly correlated edges at p<0.05 (two-tailed, FWE corrected) = 1 (minimum corrected p = 0.0166)
Number of significantly correlated edges at p<0.05 (two-tailed, uncorrected) = 12 (15 expected by chance)
Image shows edges (node-pairs) whose connection most strongly correlates with FamHist_Moth_Dep (in decreasing order), with t-statistic listed at the top of each node-pair.